Evolutionary dynamics on graphs
by Christoph Hauert, Version 1.0, December 2004.
- Location:
- VirtualLabs
- » Evolution on graphs
Evolutionary dynamics act on populations. Neither genes, nor cells, nor individuals but populations evolve. In small populations, random drift dominates, whereas large populations are sensitive to subtle differences in selective values. Traditionally, evolutionary dynamics was studied in the context of homogeneous or spatially extended populations. Here we generalize population structure by arranging individuals on a graph. Each vertex represents an individual. The fitness of an individual denotes its reproductive rate which determines how often offspring is placed into adjacent vertices. A homogenous population corresponds to a fully connected graph and spatial structures are represented by lattices where each node is connected to its nearest neighbors.
The dynamics of selection and random drift is traditionally studied by the Moran process: (a) draw a focal individual at random with a probability proportional to its fitness; (b) duplicate focal individual, i.e. create clonal offspring; and (c) replace random neighbor of focal individual with offspring. This process has the nice feature that the population size is preserved.
In the following, we study the simplest possible question: what is the probability that a single mutant generates a lineage that takes over the entire population? This fixation probability determines the rate of evolution. The higher the correlation between the mutant's fitness and its probability of fixation, the stronger the effect of natural selection; if fixation is largely independent of fitness, drift dominates.
A large class of graphs exhibit a characteristic balance between selection and drift as characterised by the isothermal theorem. Nevertheless, the structure of the graphs can have tremenduous effects on the fixation probability of mutant ranging from complete suppression of selection to complete suppression of random drift, i.e. amplification of selection. If, in addition, individuals interact with their neighbors, reflecting frequency dependent fitness, the dynamics becomes very complicated but first studies show very interesting and counterintuitive results.
This tutorial complements and illustrates a research article co-authored with Erez Lieberman and Martin Nowak in Nature (2005).
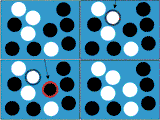 |
Static fitnessFixation probability of a single mutant in a resident population where mutants have a fixed fitness r and residents have fitness 1. Individuals do not interact and therefore the fixation probability is entirely determined by the structure of the graph. |
|---|---|
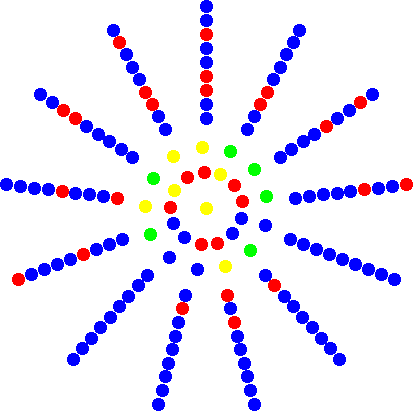 |
Frequency dependent fitnessFixation probability of a single mutant in a resident population where the fitness of both mutants and residents is determined through interactions in a local neighborhood. |
|
|
Moran process
This so-called Moran process describes the stochastic evolution in a finite population of constant size. Suppose all resident individuals are identical and one new mutant is introduced. The new mutant has relative fitness r, as compared to the residents, whose fitness is 1. The fixation probability of the mutant is then given by: R1 = (1-1/r)/(1-1/rN).
The Moran process has two absorbing states: either the population consists of all residents or all mutants. No other stable equilibrium state is possible. Whenever an absorbing state is reached, mutants (residents) are said to have reached fixation. This represents a specific balance between selection and drift: advantageous mutations have a certain chance - but no guarantee - of fixation, whereas disadvantageous mutants are likely - but again, no guarantee - to become extinct. | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
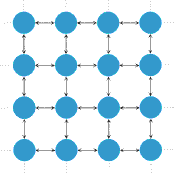
Isothermal theoremPopulation structure can be introduced by assuming that the individuals occupy the nodes of a graph. The adjacency matrix W = [wij] then determines the structure of the graph, where wij denotes the probability that individual i places its offspring into node j. If wij = wji = 0 then the nodes i and j are not connected. The temperature of a node is given by the sum over the weights of all incoming links and indicates how often it is replaced, i.e. a hot node is replaced often and a cold node is rarely replaced. The isothermal theorem now states that all isothermal graphs have fixation probability R1 if and only if all nodes have the same temperature. Isothermality is equivalent to requiring that W is doubly stochastic. All regular lattices are examples of isothermal graphs (e.g. square lattices as shown in (i) to the left). | ||||||||||||||||||
Evolutionary suppressorsThe characteristic balance between selection and drift in isothermal graphs can tilt to either side for graphs that are not isothermal. For example, suppose N individuals are arranged in a linear chain (see (i) to the left). Each individual places its offspring into the position immediately to its right. The leftmost individual is never replaced. Clearly, the fixation probability of a randomly placed mutant is 1/N, irrespective of r. The mutant can only reach fixation if it arises in the leftmost position, which happens with probability 1/N. This chain is an example of a simple population structure whose behaviour is dominated by random drift. Another example is a central hub that is connected to the nodes along the periphery but the peripherial nodes have no outgoing links (see (ii) to the left). More generally, any graph with a single root or small upstream population that feeds into large downstream populations acts as a suppressor of selection. | ||||||||||||||||||
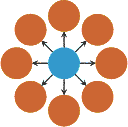
Evolutionary amplifiersIntriguingly, it is also possible to create population structures that amplify selection and suppress random drift. For example, on the star structure, where all nodes are connected to a central hub and vice versa (see (i) to the left), the fixation probability of a randomly placed mutant becomes R2 = (1-1/r)/(1-1/r2 N)
for large N. Thus, any selective difference r is amplified to r2. The super-star (see (ii) to the left for K = 3) and related structures have the amazing property that for large N, the fixation probability of any advantageous mutant converges to one, while the fixation probability of any disadvantageous mutant converges to zero. Hence, these population structures guarantee fixation of advantageous mutants however small their selective advantage. In general, we can prove that for sufficiently large population size N, a super-star of parameter K satisfies: RK = (1-1/r)/(1-1/rK N)
where K denotes the maximum number of edges connecting any two nodes, e.g. K = 1 for a fully connected graph, K = 2 for a star (any two nodes along the periphery are separated by two edges). Just as one-rooted graphs entirely suppress the effects of selection, super-stars function as arbitrarily strong amplifiers of selection and suppressors of random drift. However, note that amplification has it's price in that the average fixation time goes to infinity as the amplification increases. | ||||||||||||||||||
|
Games on graphsSo far we assumed that the individuals had a static fitness of 1 and r for residents and mutants, repsectively. However, in many situations it might be more realistic to assume frequency dependent fitness, i.e., scenarios where the fitness of an individual is not determined by its 'genotype' (mutant versus resident) but rather through interactions with other members in the population. Consider, as before, two types A and B, but instead of constant fitness, their fitness now depends on the outcome of a game with a payoff matrix as shown in the table to the left. Such interactions are often called 2×2 games (two players and two strategic types). The ranking of the payoffs a, b, c, and d determines the characteristics of the game. For example, c > a > d > b defines the famous Prisoner's Dilemma if A corresponds to cooperation and B to defection. In traditional evolutionary game dynamics, a mutant strategy A can invade a resident B if b > d. For games on graphs, the crucial condition for A invading B, and hence the very notion of evolutionary stability, can be quite different. In particular, isothermal graphs no longer have identical fixation probabilities. Unfortunately, frequency dependent selection is far more complicated because the dynamics and fixation probabilities depend not only on the graph but also on type of interactions and, moreover, it is now possible to distinguish two graphs: the replacement graph (as before) and in addition the interaction graph. At this point we have only preliminary results of this intriguing field for future research. For example, super-stars again possess powerful amplifying properties. If the payoff of an individual is determined by its downstream neighbor, the fixation probability for large N of a single A mutant is given by RK (see above) with r = (b/d)(b/c)K-1. Interestinlgy, the performance of mutants is unaffected by the performance of mutant-mutant interactions. For a super-star with large K, this r value diverges as long as b > c. Thus, even a dominated strategy (a < c and b < d) satisfying b > c will expand from a single mutant to conquer the entire super-star with a probability that can be made arbitrarily close to 1. The guaranteed fixation of this broad class of dominated strategies is a unique feature of evolutionary game theory on graphs: without structure, all dominated strategies die out. Other counter-intuitive results seem to hold for the superstar in other orientations. | |||||||||||||||||
This work was first published in
Lieberman, E., Hauert,
Ch. & Nowak, M. (2005) Evolutionary Dynamics on Graphs. Nature 433, 312-316.
Press & News
- Form follows function: the architecture of complex networks (2006) Molecular Systems Biology 2 42 by Guimerà, R. & Sales-Pardo, M.
Acknowledgements
Financial support of the Swiss National Science Foundation is gratefully acknowledged.